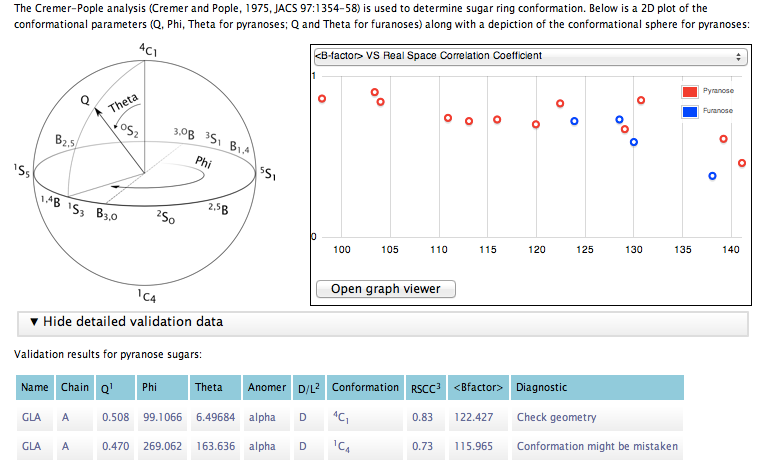
We use the well-stablished Cremer-Pople algorithm (Cremer and Pople, 1975, JACS 97:1354-58) at the core of our conformational analysis. This analysis gives one angle (Phi) and a total puckering amplitude (Q) for furanoses and two angles (Phi, Theta) and Q for pyranoses. The relationship between the different angles and conformations is reflected in the subsequent graphical representation of the conformational parameters:
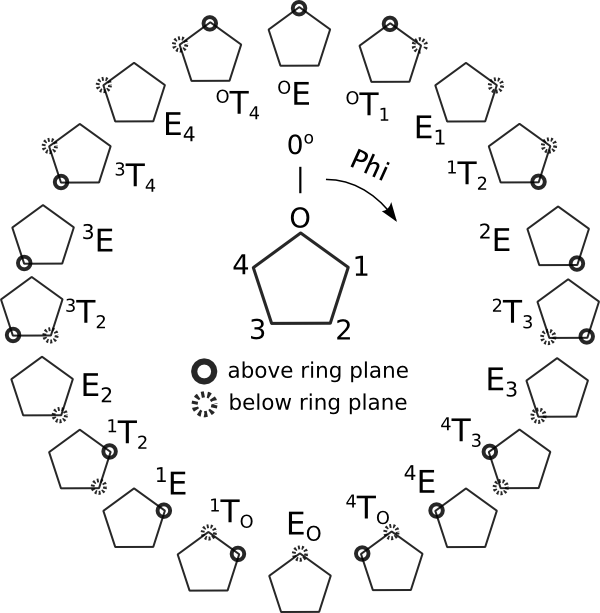
Conformational landscape for furanose sugars. Atoms without markup are roughly coplanar. This angle assignment implies a clockwise ordering of the ring atoms starting from the oxygen. An opposite-sense naming scheme would imply a 180° shift in Phi.
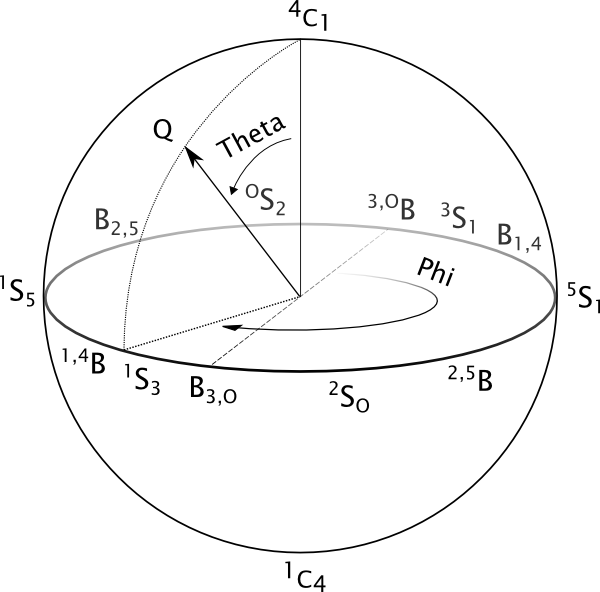
Cremer-Pople sphere for pyranoses. Chair conformations are located at the poles of the sphere, with the northern hemisphere being the one predominantly occupied by D-pyranoses (due to the configuration of the carbon linked to the highest-ranked in-ring carbon). Half-chairs and Envelopes, which have been omitted in order to produce a clearer figure, are located at Theta=45° and Theta=135° in the northern and southern hemispheres respectively.
The Cremer-Pople algorithm involves the calculation of a uniquely-defined mean plane that satisfies, among other conditions, that the sum of all the vertical distances from the ring atoms to that plane is zero. The stereochemistry can also be easily worked out by calculating equivalent displacements for those atoms relevant to it. After that, Privateer-validate compares the reported conformation and stereochemistry to those calculated for the idealised, lowest-energy structure of the studied sugar.
For a deeper nomenclature check, you can also use the pdb-care server at glycosciences.de (maintained by Dr Thomas Lütteke, Justus-Liebig University Giessen).
Approach to detection
To be completed after the publication of Privateer-detect.Program description
Privateer-validate
This validation software will analyse monosaccharide moieties from a PDB file, using a database of three-letter codes, stereochemistry and conformations derived from the wwPDB database. It will also output linear descriptions of any glycans found (N- or O-glycans). The program takes as input a set of structure factors in MTZ (amplitudes or intensities) or CIF (amplitudes only) formats and a structure in PDB format, and determines the conformation and stereochemistry for all the monosaccharides it's able to find in the structure file. Additionally, a Real Space Correlation coefficient is computed against an omit map calculated excluding the sugars from phase calculations. The program will also produce a visual checklist with the conflicting sugar models in the form of Scheme and Python scripts for use with Coot.To use them:
coot --script privateer-results.py or coot --script privateer-results.scmBlue map: 2mFo-DFc map. Pink map: mFo-DFc omit map.
Running Coot with these scripts also automatically turns on torsion restraints for real space refinement. This means that there is a better chance of keeping the sugars in their original low-energy conformation if they have not deviated too much from it.
Privateer-validate offers an easy-to-use graphical interface that runs under CCP4i2, and which produces richer reports than those available from its text-based version. The input data window looks like this:
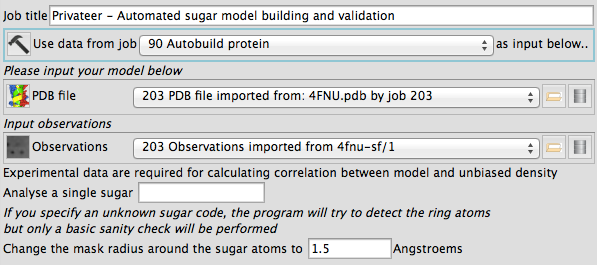
Here is a look at the report window for a dataset where furanoses and pyranoses coexist:
And finally, here is an interactive piece on the glycosylation tree structure reports. You may place your mouse over a sugar to reveal the three-letter code and conformation:
Man←α6−Man←β4−GlcNac← β4−GlcNac ← β−ASN61
GlcNac ← β−ASN211
Man←β4−GlcNac←β4−GlcNac ← β−ASN252
GlcNac←β4−GlcNac ← β−ASN315
Man←α2−Man←α6−(Man←α2−Man←α3−)Man←α6−(Man←α2−Man←α3−)Man←β4−GlcNac←β4−GlcNac ← β−ASN322
If you choose to rebuild your structure with Coot after running Privateer, a visual guided tour of the reported issues will be automatically loaded. Torsion restraints will also be automatically turned on.
Command line usage
prompt% privateer-validate -pdbin file.pdb -mtzin file.mtz -colin-fo F_i24,SIGF_i24Parameter list
-pdbin [file.pdb]- Compulsory argument: input PDB file to examine. If no CRYST1 card is present, Privateer will still process it but please bear in mind that it may be missing important contacts described by crystallographic symmetry.
-mtzin [file.mtz]or-cifin [file.cif]- An MTZ file containing amplitudes or intensities or a CIF file containing amplitudes. If intensities are supplied, Privateer will run the CCP4 program 'ctruncate' in order to produce amplitudes. Please ensure that CCP4 software is within reach (i.e. included in the PATH variable). Privateer-validate is compatible out of the box with most of the CIF structure factor files downloaded from the PDB. Compatible with miniMTZ files from CCP4i2.
-colin-fo [F,SIGF]- Columns containing F and SIGF, e.g. FOBS,SIGFOBS or F,SIGF without any spaces in between characters. If not supplied, Privateer will try to guess the path using the most commonly used choices.
-codein [SUG]- Three-letter code (those defined by the wwPDB) for the target sugar. If this parameter is not provided, Privateer will scan and score all monosaccharides present in it's database.
-showgeom- Bond lengths, angles and torsions are reported for each ring.
-mtzout [file.mtz]- Optional: output map coefficients to an MTZ file. A name for the file must be provided.
-radiusin [value in angstroems]- Optional: provide a radius for the calculation of the mask around the target sugar. Default value is 1.5 Angstroems.
-mode [normal|ccp4i2]- Optional: run mode. Default is normal. Under ccp4i2 mode, Privateer will generate miniMTZ files, less text output and XML data containing HTML descriptions of any glycosylation trees found, and all the validation data.
Privateer
To be published very soon.Source code
Privateer is Free Software. You can access and/or checkout it's source code at CCP4forge (Bazaar repository, stable versions) or Google Code repository (Subversion repository, experimental stuff that may not work).Authors
Jon Agirre and Kevin CowtanYork Structural Biology Laboratory
Department of Chemistry, The University of York
Wentworth Way, YO10 5DD, England.
Last modified: December 1st, 2014.
Bugs can be reported to jon.agirre (at) york.ac.uk
Acknowledgements
A number of people have shaped the future of Privateer with their ideas. In no particular order: Eleanor Dodson, Jacob Sřrensen, Keith Wilson, Gideon Davies, Christian 'Bones' Roth, Wendy Offen, Thomas Lütteke, Carme Rovira, Jose A. Cuesta Seijo, Stuart McNicholas, Antoni Planas, Marcelo Guerin, Dan Wright, Glyn Hemsworth, Andrew Thompson, Marcin Wojdyr, Saioa Urresti, Paul Emsley and Robbie Joosten.[Back to top]